How is resolution computed
Index
[1]:
import numpy as np
import matplotlib.pyplot as plt
from ctaplot.ana import resolution, relative_scaling
Normal distribution
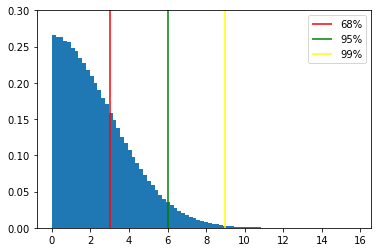
 corresponds to the 68 percentile of the distribution
corresponds to the 68 percentile of the distribution[2]:
loc = 10
scale = 3
X = np.random.normal(size=1000000, scale=scale, loc=loc)
plt.hist(np.abs(X - loc), bins=80, density=True)
sig_68 = np.percentile(np.abs(X - loc), 68.27)
sig_95 = np.percentile(np.abs(X - loc), 95.45)
sig_99 = np.percentile(np.abs(X - loc), 99.73)
plt.vlines(sig_68, 0, 0.3, label='68%', color='red')
plt.vlines(sig_95, 0, 0.3, label='95%', color='green')
plt.vlines(sig_99, 0, 0.3, label='99%', color='yellow')
plt.ylim(0,0.3)
plt.legend()
print("68th percentile = {:.4f}".format(sig_68))
print("95th percentile = {:.4f}".format(sig_95))
print("99th percentile = {:.4f}".format(sig_99))
assert np.isclose(sig_68, scale, rtol=1e-2)
assert np.isclose(sig_95, 2 * scale, rtol=1e-2)
assert np.isclose(sig_99, 3 * scale, rtol=1e-2)
68th percentile = 2.9999
95th percentile = 6.0005
99th percentile = 8.9907
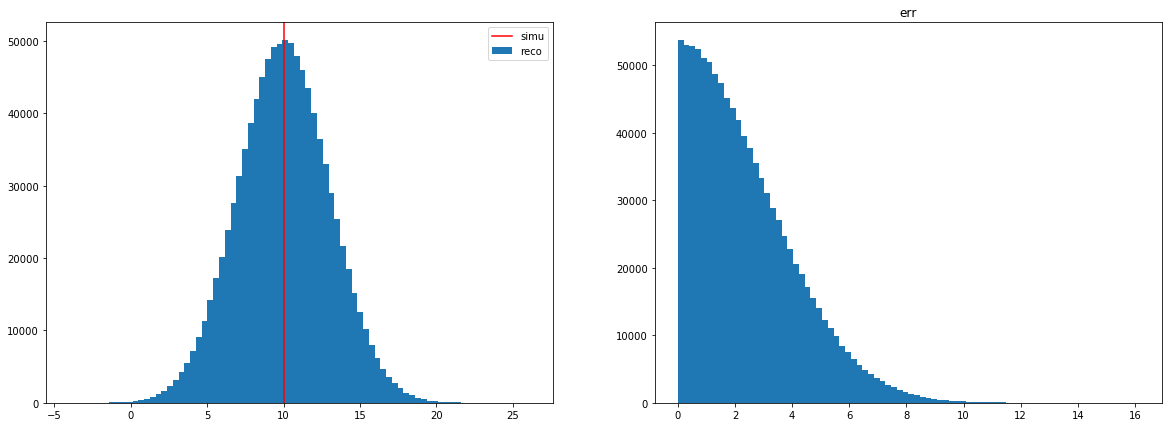
Resolution
err = (reco - simu)/scaling.[3]:
simu = loc * np.ones(X.shape[0])
reco = np.random.normal(loc=loc, scale=scale, size=X.shape[0])
err = np.abs(simu - reco)
fig, axes = plt.subplots(1, 2, figsize=(20,7))
axes[0].hist(reco, bins=80, label='reco')
axes[0].axvline(loc, 0, 1, color='red', label='simu')
axes[0].legend()
axes[1].hist(err, bins=80)
axes[1].set_title('err')
[3]:
Text(0.5, 1.0, 'err')
reco in order to have a relative error equals to err.[4]:
res = resolution(simu, reco, relative_scaling_method=None)
print(res)
assert np.isclose(res[0], scale, rtol=1e-2)
[3.0029721 2.99821785 3.00797764]
Relative scaling
The resolution can be measured on the absolute error or on the relative one.

ctaplot.relative_scaling_method:s0 : no scaling (scaling = 1)s1 : $ scaling = |simu| $s2 : $ scaling = |reco| $s3 : $ scaling = (|simu\| + \|reco|)/2 $s4 : $ scaling = max(|reco|, |simu|) $The default one for the resolution is s1.
Note that methods involving reco are more subject to deviation from the expected result:
[5]:
for method in ['s1', 's2', 's3', 's4']:
res = resolution(simu, reco, relative_scaling_method=method)
print("Method {} gives a resolution = {:.3f} to be compared with the expected value = {}".format(method, res[0], scale/loc))
Method s1 gives a resolution = 0.300 to be compared with the expected value = 0.3
Method s2 gives a resolution = 0.288 to be compared with the expected value = 0.3
Method s3 gives a resolution = 0.297 to be compared with the expected value = 0.3
Method s4 gives a resolution = 0.259 to be compared with the expected value = 0.3
s1Error bars
The implementation for percentile confidence interval follows: - http://people.stat.sfu.ca/~cschwarz/Stat-650/Notes/PDF/ChapterPercentiles.pdf
Example with a normal distribution:
[6]:
nbins, bins, patches = plt.hist(X, bins=100, density=True)
ymax = 1.1 * nbins.max()
plt.ylim(0, ymax)
plt.vlines(np.percentile(X, 68.27), 0, ymax, color='red')
plt.show()
print("The true 68th percentile of this distribution is: {:.4f}".format(np.percentile(X, 68.27)))
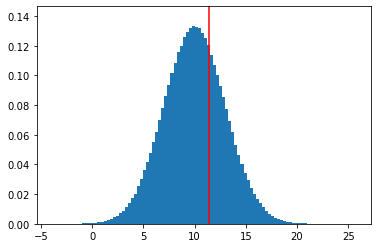
The true 68th percentile of this distribution is: 11.4226
[7]:
n = 1000
p = 0.6827
all_68 = []
for i in range(int(len(X)/n)):
all_68.append(np.percentile(X[i*n:(i+1)*n], p*100))
all_68 = np.array(all_68)
nbins, bins, patches = plt.hist(all_68, density=True);
ymax = 1.1 * nbins.max()
plt.vlines(all_68.mean(), 0, ymax)
plt.vlines(all_68.mean() + all_68.std(), 0, ymax, color='red')
plt.vlines(all_68.mean() - all_68.std(), 0, ymax, color='red')
plt.ylim(0, ymax)
plt.title("Distribution of the measured 68th percentile")
print("Standard deviation = {:.5f}".format(all_68.std()))
Standard deviation = 0.11973
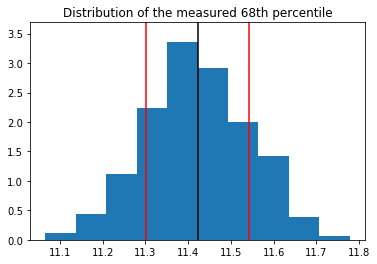
To evaluate directly the confidence interval from a sub-sample of the distribution, one can use the following formulae:
$ R_{low} = np − z \sqrt{n * p * (1 − p)} $
$ R_{up} = np + z \sqrt{n * p * (1 − p)} $
with  the percentile and
the percentile and  the confidence level desired.
the confidence level desired.
And the confidence interval given by: (![X[R_{low}], X[R_{up}])](_images/math/2ee978c036ec10438ce6737db61f0279a9f7fc40.png)
scipy.stats.norm.ppf).z = 0.47 for a confidence level of 68%
z = 1.645 for a confidence level of 95%
z = 2.33 for a confidence level of 99%
[8]:
# confidence level:
z = 2.33
# sub-sample:
x = X[:n]
rl = int(n * p - z * np.sqrt(n * p * (1-p)))
ru = int(n * p + z * np.sqrt(n * p * (1-p)))
print("Measured percentile: {:.4f}".format(np.percentile(x, p*100)))
print("Confidence interval: ({:.4f}, {:.4f})".format(np.sort(x)[rl], np.sort(x)[ru]))
print("To be compared with: ({:.4f}, {:.4f})".format(all_68.mean()-all_68.std()*3, all_68.mean()+all_68.std()*3))
print("which is the corresponding confidence interval given by the normal distribution of the measured percentiles")
Measured percentile: 11.5283
Confidence interval: (11.2443, 11.9176)
To be compared with: (11.0628, 11.7812)
which is the corresponding confidence interval given by the normal distribution of the measured percentiles
In ctaplot, this is computed by the function percentile_confidence interval:
[9]:
from ctaplot.ana import percentile_confidence_interval
p = 68.27
confidence_level = 0.99
pci = percentile_confidence_interval(x, percentile=p, confidence_level=0.99)
print("68th percentile: {:.3f}".format(np.percentile(x, p)))
print("Interval with a confidence level of {}%: ({:.3f}, {:.3f})".format(confidence_level*100, pci[0], pci[1]))
plt.figure(figsize=(12,7))
nbins, bins, patches = plt.hist(x, bins=100, density=True);
ymax = 1.1 * nbins.max()
plt.vlines(np.percentile(x, 50), 0, ymax, color='black')
plt.vlines(np.percentile(x, p), 0, ymax, color='red')
plt.vlines(pci[0], 0, ymax, linestyles='--', color='red')
plt.vlines(pci[1], 0, ymax, linestyles='--', color='red',)
plt.ylim(0, ymax);
68th percentile: 11.528
Interval with a confidence level of 99.0%: (11.244, 11.918)
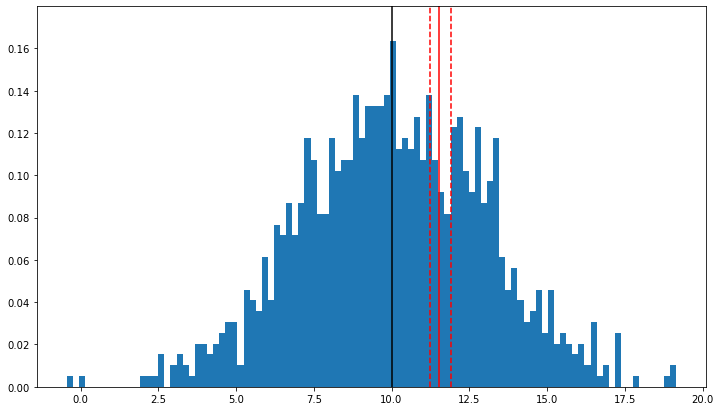
 for a normal distribution.
for a normal distribution.[10]:
pci = percentile_confidence_interval(X, percentile=50, confidence_level=0.99)
print("Median: {:.5f}".format(np.median(X)))
print("Confidence interval at 99%: {}".format(pci))
print("To be compared with: ({}, {})".format(np.median(X)-3*scale/np.sqrt(len(X)), np.median(X)+3*scale/np.sqrt(len(X))))
Median: 9.99912
Confidence interval at 99%: (9.990545100476202, 10.007734065899768)
To be compared with: (9.990119056062714, 10.008119056062714)